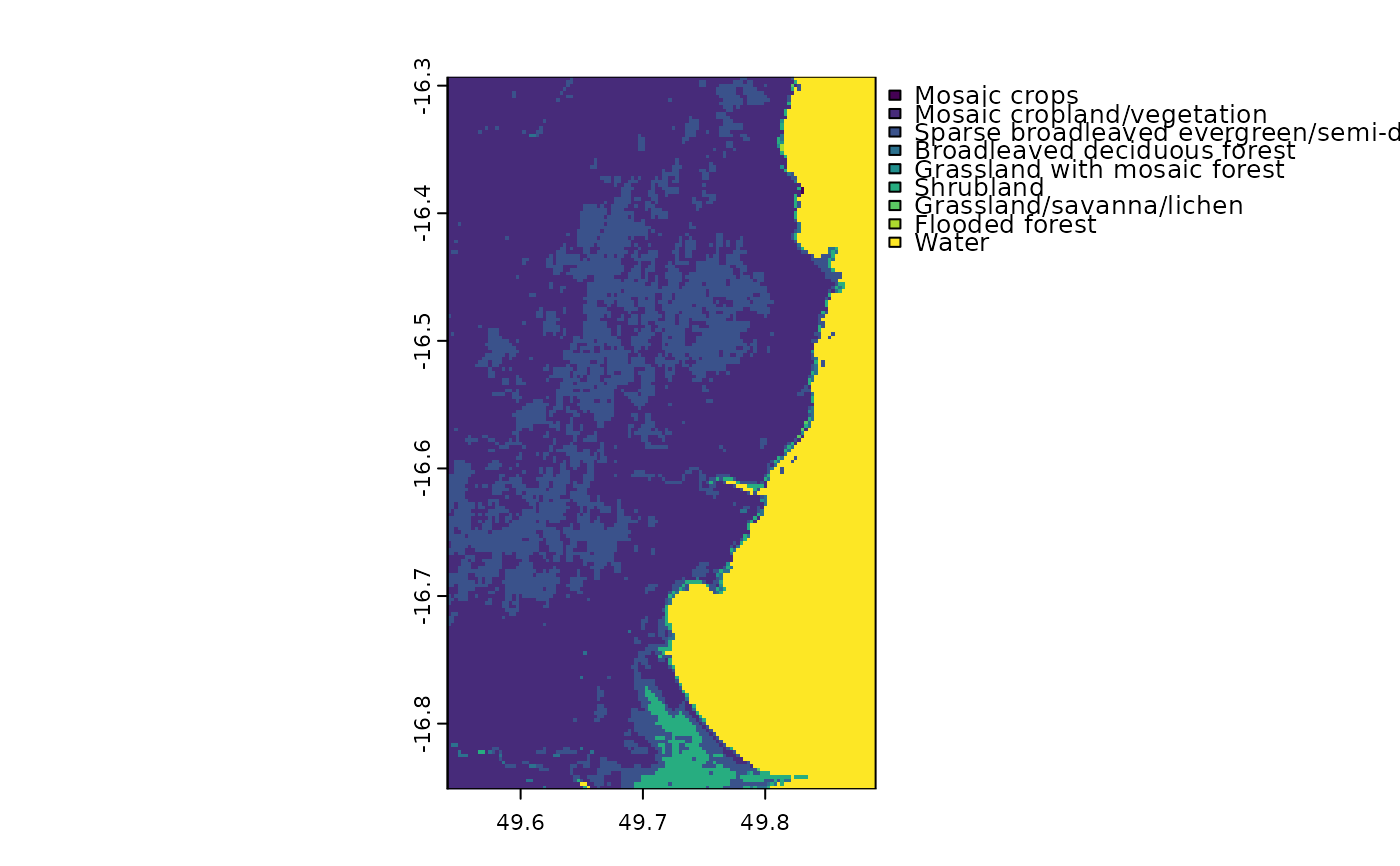
Raster of land cover for an eastern portion of Madagascar. Note that the land cover classes have been simplified, so this raster should not be used for "real" analyses.
References
Arino O., P. Bicheron, F. Achard, J. Latham, R. Witt and J.-L. Weber. 2008. GlobCover: The most detailed portrait of Earth. European Space Agency Bulletin 136:25-31. http://due.esrin.esa.int.
Examples
### vector data
library(sf)
# For vector data, we can use data(*) or fastData(*):
data(madCoast0) # same as next line
madCoast0 <- fastData("madCoast0") # same as previous
madCoast0
#> Simple feature collection with 1 feature and 68 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: 731581.6 ymin: 1024473 xmax: 768721.2 ymax: 1085686
#> Projected CRS: Tananarive (Paris) / Laborde Grid
#> OBJECTID ID_0 ISO NAME_ENGLISH NAME_ISO NAME_FAO NAME_LOCAL NAME_OBSOLETE
#> 1 1 134 MDG Madagascar MADAGASCAR Madagascar Madagascar
#> NAME_VARIANTS NAME_NONLATIN NAME_FRENCH NAME_SPANISH NAME_RUSSIAN
#> 1 Malagasy Republic Madagascar Madagascar Мадагаскар
#> NAME_ARABIC NAME_CHINESE WASPARTOF CONTAINS SOVEREIGN ISO2 WWW FIPS ISON
#> 1 مدغشقر 马达加斯加 Madagascar MG MA 450
#> VALIDFR VALIDTO POP2000 SQKM POPSQKM UNREGION1
#> 1 19581014 Present 15970364 594856.375 26.847428507427527 Eastern Africa
#> UNREGION2 DEVELOPING CIS Transition OECD WBREGION WBINCOME
#> 1 Africa 1 Sub-Saharan Africa Low income
#> WBDEBT WBOTHER CEEAC CEMAC CEPLG COMESA EAC ECOWAS IGAD IOC MRU
#> 1 Moderately indebted HIPC 1 1
#> SACU UEMOA UMA PALOP PARTA CACM EurAsEC Agadir SAARC ASEAN NAFTA GCC CSN
#> 1
#> CARICOM EU CAN ACP Landlocked AOSIS SIDS Islands LDC
#> 1 1 1 1
#> geometry
#> 1 MULTIPOLYGON (((755432.2 10...
plot(st_geometry(madCoast0))
madCoast4 <- fastData("madCoast4")
madCoast4
#> Simple feature collection with 2 features and 17 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: 731811.7 ymin: 1024542 xmax: 768726.5 ymax: 1085485
#> Projected CRS: Tananarive (Paris) / Laborde Grid
#> OBJECTID ID_0 ISO NAME_0 ID_1 NAME_1 ID_2 NAME_2 ID_3
#> 1 1070 134 MDG Madagascar 5 Toamasina 17 Analanjirofo 79
#> 2 1098 134 MDG Madagascar 5 Toamasina 17 Analanjirofo 82
#> NAME_3 ID_4 NAME_4 VARNAME_4 CCN_4 CCA_4 TYPE_4 ENGTYPE_4
#> 1 Mananara 1070 Antanambe NA Fokontany Commune
#> 2 Soanierana-Ivongo 1098 Manompana NA Fokontany Commune
#> geometry
#> 1 MULTIPOLYGON (((760305.9 10...
#> 2 MULTIPOLYGON (((754786.8 10...
plot(st_geometry(madCoast4), add = TRUE)
madRivers <- fastData("madRivers")
madRivers
#> Simple feature collection with 11 features and 5 fields
#> Geometry type: LINESTRING
#> Dimension: XY
#> Bounding box: xmin: 731627.1 ymin: 1024541 xmax: 762990.1 ymax: 1085580
#> Projected CRS: Tananarive (Paris) / Laborde Grid
#> First 10 features:
#> F_CODE_DES HYC_DESCRI NAM ISO NAME_0
#> 1 River/Stream Perennial/Permanent MANANARA MDG Madagascar
#> 2 River/Stream Perennial/Permanent MANANARA MDG Madagascar
#> 3 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 4 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 5 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 6 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 7 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 8 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 9 River/Stream Perennial/Permanent UNK MDG Madagascar
#> 10 River/Stream Perennial/Permanent UNK MDG Madagascar
#> geometry
#> 1 LINESTRING (739818.2 108005...
#> 2 LINESTRING (739818.2 108005...
#> 3 LINESTRING (747857.8 108558...
#> 4 LINESTRING (739818.2 108005...
#> 5 LINESTRING (762990.1 105737...
#> 6 LINESTRING (742334.2 106858...
#> 7 LINESTRING (731803.7 105391...
#> 8 LINESTRING (755911.6 104957...
#> 9 LINESTRING (731871 1044531,...
#> 10 LINESTRING (750186.1 103441...
plot(st_geometry(madRivers), col = "blue", add = TRUE)
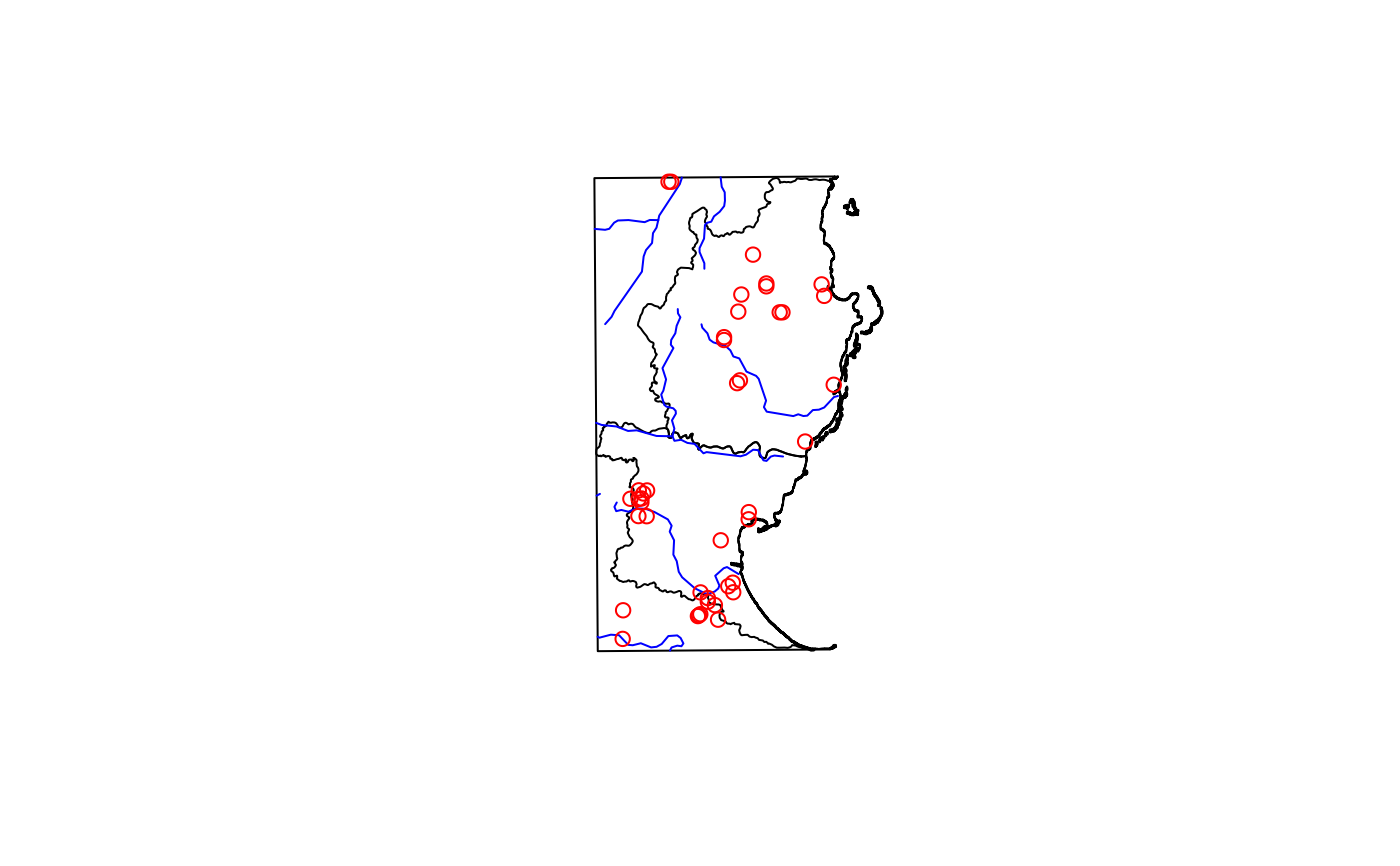
madDypsis <- fastData("madDypsis")
madDypsis
#> Simple feature collection with 42 features and 9 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 735228.4 ymin: 1026056 xmax: 762442 ymax: 1085002
#> Projected CRS: Tananarive (Paris) / Laborde Grid
#> First 10 features:
#> gbifID license rightsHolder institutionCode year
#> 1 2397516155 CC_BY_NC_4_0 vononarbgkew iNaturalist 2019
#> 2 2397516017 CC_BY_NC_4_0 vononarbgkew iNaturalist 2019
#> 3 2397515145 CC_BY_NC_4_0 vononarbgkew iNaturalist 2019
#> 4 2268865622 CC_BY_4_0 Missouri Botanical Garden MO 2006
#> 5 2268863965 CC_BY_4_0 Missouri Botanical Garden MO 1991
#> 6 2268862328 CC_BY_4_0 Missouri Botanical Garden MO 1994
#> 7 2268862230 CC_BY_4_0 Missouri Botanical Garden MO 1991
#> 8 1928075921 CC0_1_0 The New York Botanical Garden NY 2006
#> 9 1677261542 CC_BY_NC_4_0 Landy Rita iNaturalist 2016
#> 10 1453257920 CC_BY_NC_4_0 mamy_andriamahay iNaturalist 2016
#> month day coordinateUncertaintyInMeters species
#> 1 8 11 4 Dypsis nodifera
#> 2 8 11 3 Dypsis nodifera
#> 3 8 11 3 Dypsis nodifera
#> 4 9 14 NA Dypsis betsimisarakae
#> 5 10 10 NA Dypsis nodifera
#> 6 10 23 NA Dypsis nodifera
#> 7 10 11 NA Dypsis nodifera
#> 8 9 15 NA Dypsis integra
#> 9 6 11 17 Dypsis lastelliana
#> 10 11 29 21 Dypsis lastelliana
#> geometry
#> 1 POINT (744929.8 1028994)
#> 2 POINT (745240.1 1029239)
#> 3 POINT (745067.4 1029098)
#> 4 POINT (737649.4 1044160)
#> 5 POINT (760879.5 1071766)
#> 6 POINT (748297.4 1064593)
#> 7 POINT (747876.6 1038768)
#> 8 POINT (737901.5 1044806)
#> 9 POINT (749428.6 1033303)
#> 10 POINT (745272.7 1032050)
plot(st_geometry(madDypsis), col = "red", add = TRUE)
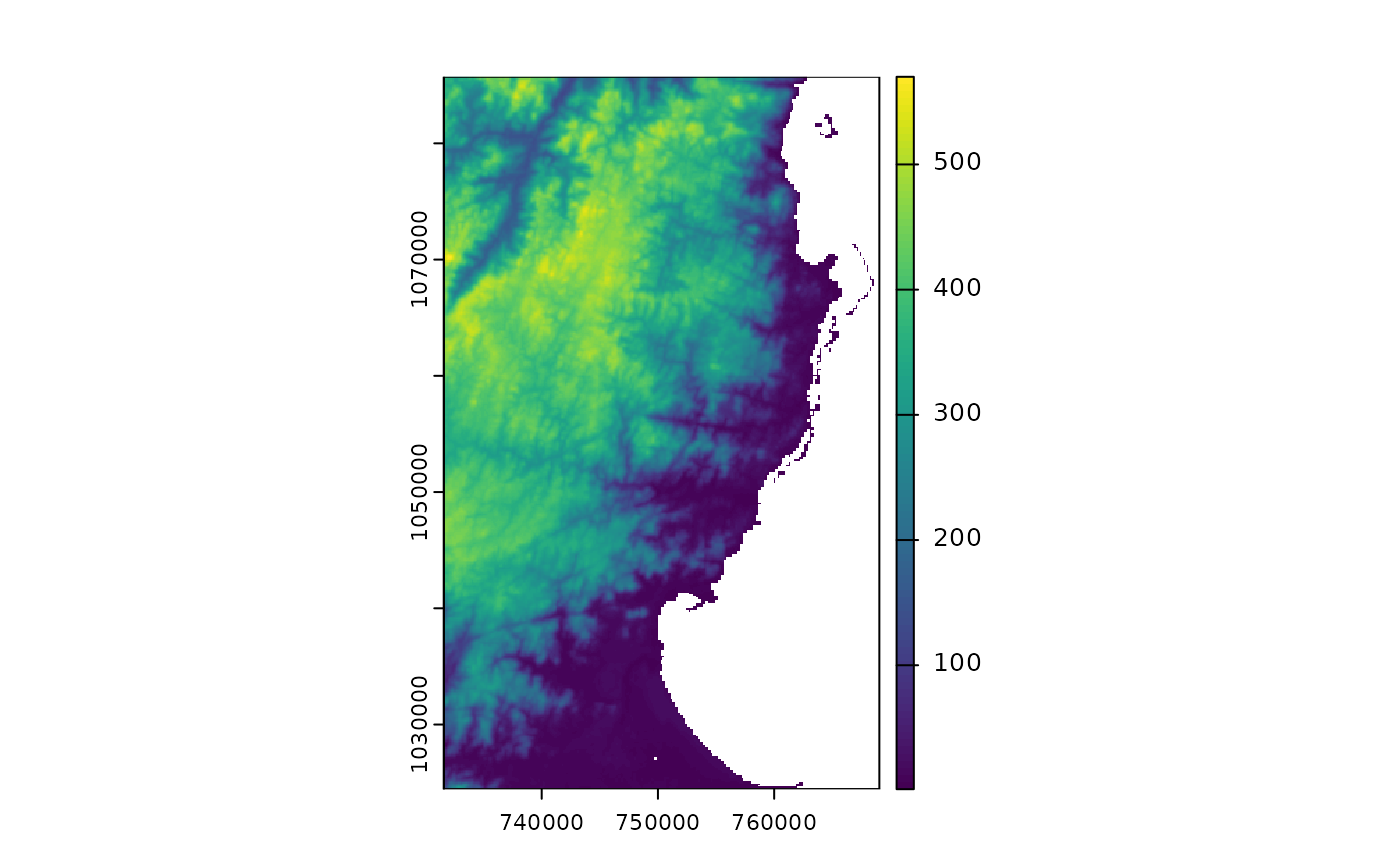
 ### raster data
library(terra)
# For raster data, we can get the file directly or using fastData(*):
rastFile <- system.file("extdata/madElev.tif", package="fasterRaster")
madElev <- terra::rast(rastFile)
madElev <- fastData("madElev") # same as previous two lines
madElev
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madElev.tif
#> name : madElev
#> min value : 1
#> max value : 570
plot(madElev)
### raster data
library(terra)
# For raster data, we can get the file directly or using fastData(*):
rastFile <- system.file("extdata/madElev.tif", package="fasterRaster")
madElev <- terra::rast(rastFile)
madElev <- fastData("madElev") # same as previous two lines
madElev
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madElev.tif
#> name : madElev
#> min value : 1
#> max value : 570
plot(madElev)
 madForest2000 <- fastData("madForest2000")
madForest2000
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madForest2000.tif
#> name : madForest2000
#> min value : 1
#> max value : 1
plot(madForest2000)
madForest2000 <- fastData("madForest2000")
madForest2000
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madForest2000.tif
#> name : madForest2000
#> min value : 1
#> max value : 1
plot(madForest2000)
 madForest2014 <- fastData("madForest2014")
madForest2014
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madForest2014.tif
#> name : madForest2014
#> min value : 1
#> max value : 1
plot(madForest2014)
madForest2014 <- fastData("madForest2014")
madForest2014
#> class : SpatRaster
#> size : 512, 313, 1 (nrow, ncol, nlyr)
#> resolution : 119.7031, 119.7031 (x, y)
#> extent : 731581.6, 769048.6, 1024437, 1085725 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madForest2014.tif
#> name : madForest2014
#> min value : 1
#> max value : 1
plot(madForest2014)
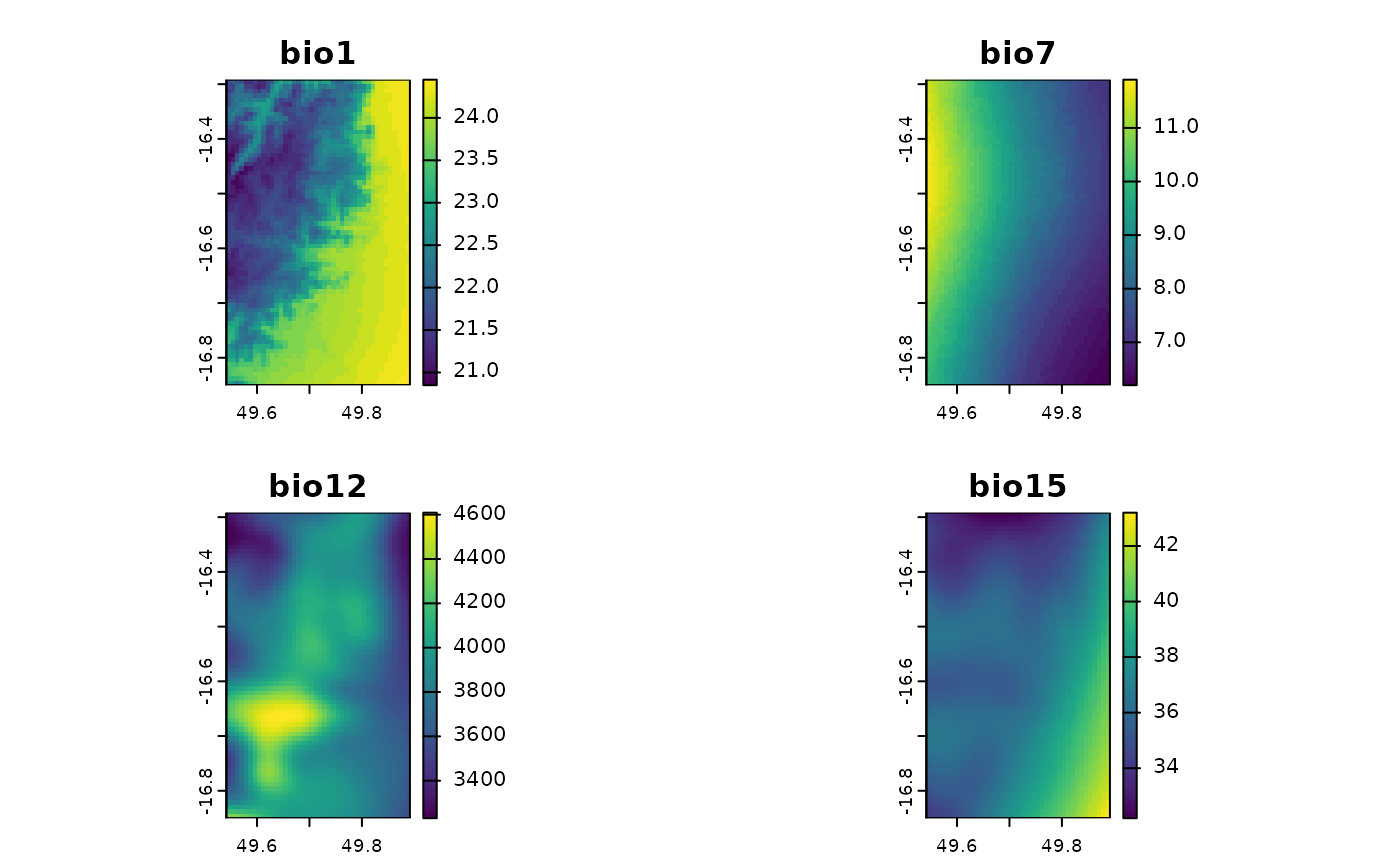
 # multi-layer rasters
madChelsa <- fastData("madChelsa")
madChelsa
#> class : SpatRaster
#> size : 67, 42, 4 (nrow, ncol, nlyr)
#> resolution : 0.008333333, 0.008333333 (x, y)
#> extent : 49.54153, 49.89153, -16.85014, -16.29181 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source : madChelsa.tif
#> names : bio1, bio7, bio12, bio15
#> min values : 20.85, 6.2, 3230.9, 32.2
#> max values : 24.45, 11.9, 4608.9, 43.2
plot(madChelsa)
# multi-layer rasters
madChelsa <- fastData("madChelsa")
madChelsa
#> class : SpatRaster
#> size : 67, 42, 4 (nrow, ncol, nlyr)
#> resolution : 0.008333333, 0.008333333 (x, y)
#> extent : 49.54153, 49.89153, -16.85014, -16.29181 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source : madChelsa.tif
#> names : bio1, bio7, bio12, bio15
#> min values : 20.85, 6.2, 3230.9, 32.2
#> max values : 24.45, 11.9, 4608.9, 43.2
plot(madChelsa)
 madPpt <- fastData("madPpt")
madTmin <- fastData("madTmin")
madTmax <- fastData("madTmax")
madPpt
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madPpt.tif
#> names : ppt01, ppt02, ppt03, ppt04, ppt05, ppt06, ...
#> min values : 344, 343, 343, 246, 146, 161, ...
#> max values : 379, 401, 442, 395, 265, 261, ...
madTmin
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madTmin.tif
#> names : tmin01, tmin02, tmin03, tmin04, tmin05, tmin06, ...
#> min values : 21, 21, 20, 19, 17, 16, ...
#> max values : 23, 23, 23, 22, 20, 19, ...
madTmax
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madTmax.tif
#> names : tmax01, tmax02, tmax03, tmax04, tmax05, tmax06, ...
#> min values : 29, 29, 29, 28, 27, 25, ...
#> max values : 31, 31, 30, 30, 28, 27, ...
# RGB raster
madLANDSAT <- fastData("madLANDSAT")
madLANDSAT
#> class : SpatRaster
#> size : 344, 209, 4 (nrow, ncol, nlyr)
#> resolution : 180, 180 (x, y)
#> extent : 344055, 381675, -1863345, -1801425 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / UTM zone 39N (EPSG:32639)
#> source : madLANDSAT.tif
#> names : band2, band3, band4, band5
#> min values : 15, 23, 22, 25
#> max values : 157, 154, 158, 166
plotRGB(madLANDSAT, 4, 1, 2, stretch = "lin")
madPpt <- fastData("madPpt")
madTmin <- fastData("madTmin")
madTmax <- fastData("madTmax")
madPpt
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madPpt.tif
#> names : ppt01, ppt02, ppt03, ppt04, ppt05, ppt06, ...
#> min values : 344, 343, 343, 246, 146, 161, ...
#> max values : 379, 401, 442, 395, 265, 261, ...
madTmin
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madTmin.tif
#> names : tmin01, tmin02, tmin03, tmin04, tmin05, tmin06, ...
#> min values : 21, 21, 20, 19, 17, 16, ...
#> max values : 23, 23, 23, 22, 20, 19, ...
madTmax
#> class : SpatRaster
#> size : 9, 6, 12 (nrow, ncol, nlyr)
#> resolution : 6082.837, 6082.837 (x, y)
#> extent : 726346.3, 762843.4, 1026783, 1081528 (xmin, xmax, ymin, ymax)
#> coord. ref. : Tananarive (Paris) / Laborde Grid
#> source : madTmax.tif
#> names : tmax01, tmax02, tmax03, tmax04, tmax05, tmax06, ...
#> min values : 29, 29, 29, 28, 27, 25, ...
#> max values : 31, 31, 30, 30, 28, 27, ...
# RGB raster
madLANDSAT <- fastData("madLANDSAT")
madLANDSAT
#> class : SpatRaster
#> size : 344, 209, 4 (nrow, ncol, nlyr)
#> resolution : 180, 180 (x, y)
#> extent : 344055, 381675, -1863345, -1801425 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / UTM zone 39N (EPSG:32639)
#> source : madLANDSAT.tif
#> names : band2, band3, band4, band5
#> min values : 15, 23, 22, 25
#> max values : 157, 154, 158, 166
plotRGB(madLANDSAT, 4, 1, 2, stretch = "lin")
 # categorical raster
madCover <- fastData("madCover")
madCover
#> class : SpatRaster
#> size : 201, 126, 1 (nrow, ncol, nlyr)
#> resolution : 0.002777778, 0.002777778 (x, y)
#> extent : 49.54028, 49.89028, -16.85139, -16.29306 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source : madCover.tif
#> categories : Short, Long
#> name : Short
#> min value : Mosaic crops
#> max value : Water
madCover <- droplevels(madCover)
levels(madCover) # levels in the raster
#> [[1]]
#> Value Short
#> 1 20 Mosaic crops
#> 2 30 Mosaic cropland/vegetation
#> 3 40 Sparse broadleaved evergreen/semi-deciduous forest
#> 4 50 Broadleaved deciduous forest
#> 5 120 Grassland with mosaic forest
#> 6 130 Shrubland
#> 7 140 Grassland/savanna/lichen
#> 8 170 Flooded forest
#> 9 210 Water
#>
nlevels(madCover) # number of categories
#> [1] 0
catNames(madCover) # names of categories table
#> [[1]]
#> [1] "Value" "Short" "Long"
#>
plot(madCover)
# categorical raster
madCover <- fastData("madCover")
madCover
#> class : SpatRaster
#> size : 201, 126, 1 (nrow, ncol, nlyr)
#> resolution : 0.002777778, 0.002777778 (x, y)
#> extent : 49.54028, 49.89028, -16.85139, -16.29306 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (EPSG:4326)
#> source : madCover.tif
#> categories : Short, Long
#> name : Short
#> min value : Mosaic crops
#> max value : Water
madCover <- droplevels(madCover)
levels(madCover) # levels in the raster
#> [[1]]
#> Value Short
#> 1 20 Mosaic crops
#> 2 30 Mosaic cropland/vegetation
#> 3 40 Sparse broadleaved evergreen/semi-deciduous forest
#> 4 50 Broadleaved deciduous forest
#> 5 120 Grassland with mosaic forest
#> 6 130 Shrubland
#> 7 140 Grassland/savanna/lichen
#> 8 170 Flooded forest
#> 9 210 Water
#>
nlevels(madCover) # number of categories
#> [1] 0
catNames(madCover) # names of categories table
#> [[1]]
#> [1] "Value" "Short" "Long"
#>
plot(madCover)